FSHD Panel Now Includes Additional Targets, Wednesday, April 5, 2023
Effective April 5th, 2023
FSHD Panel (LAB8104) now includes additional testing for DNMT3B and LRIF1genes.
Approximately 90% of individuals affected with FSHD have a chromosome 4q35 deletion. The identification of a characteristic 4q35 deletion is more than 90% specific for the disease. Furthermore, patients with FSHD have 4qA alleles. A deleted 4q35A allele is diagnostic of FSHD type 1 (FSHD1). Approximately 5% of FSHD patients have FSHD type 2. These patients have non-deleted 4q35A alleles, hypomethylation of D4Z4 repeats, and either a heterozygous dominant mutation in SMCHD1 or DNMT3B or biallelic recessive mutations in LRIF1.
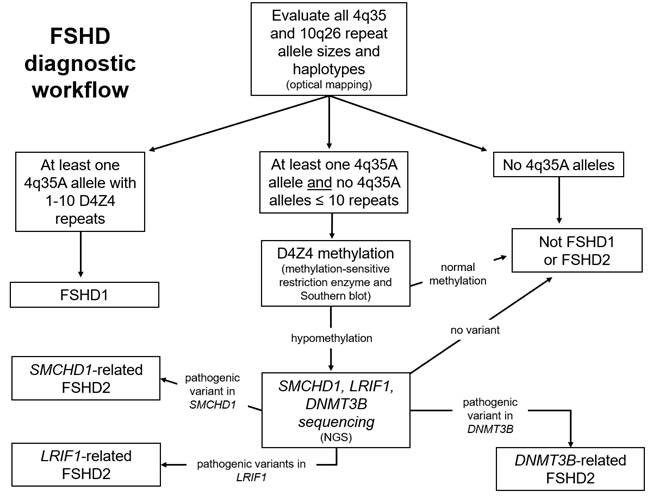
Peripheral blood leukocytes (or cultured cells) have high molecular weight DNA extracted. FSHD analysis via Bionano DLS labeling, Saphyr instrumentation, and Enfocus FSHD specific pipeline/de novo assembly pipeline is based on optical mapping data collected on the Saphyr Genome Imaging instrument. Based on specific labeling and mapping of ultra-high molecular weight DNA in nanochannel arrays, optical mapping provides a high-resolution analysis of the D4Z4 repeat array. The molecules aligning to regions of interest are extracted and assembled. The resulting consensus maps are used for the Bionano EnFocus FSHD Analysis. The D4Z4 repeat regions in chromosomes 4 and10 are sized, and the permissive and non-permissive haplotypes (4qA and 4qB) assigned. Additional structural variants and copy number gains and losses are noted in the proximity of the D4Z4 repeat array on chromosome 4 and of the SMCHD1 gene on chromosome 18.
Testing for FSHD2 involves determining the methylation status of the D4Z4 repeats, via Southern blot analysis, that uses isolated DNA. In patients with a permissive 4qA allele and an appropriate level of hypomethylation, mutation analysis of the SMCHD1, LRIF1, and DNMT3B genes is performed on a next-generation sequencing platform.
Testing requires peripheral blood collected in two dedicated pink top EDTA tubes. A minimum volume of 10 ml is recommended. Testing is performed using optical genome mapping, Southern blotting, and targeted next-generation sequencing.
During this transition period, specimens collected in the older Hemoccult ICT will still be analyzed and then resulted in Epic.
Questions can be directed to Steven Moore, MD (384-9084, steven-moore@uiowa.edu) or Sarah Hornberg, Molecular Pathology Laboratory Supervisor (384-9870, sarah-hornberg@uiowa.edu).