Facioscapulohumeral Dystrophy (FSHD) Information (FSHD1 and FSHD2)
| Blood Samples: |
Adults - whole blood in pink top tubes (EDTA) Children - whole blood in pink top tubes (EDTA) The optimum sample is 10 mL whole blood in EDTA Pink top tubes. Optical Genome Mapping (OGM) for 4q35 deletion detection, 4q/10q haplotype determination, methylation testing, and SMCHD1 gene sequencing for FSHD2 utilize isolated DNA from peripheral blood leukocytes. The minimum blood volume for testing depends on the medical circumstances and the assays requested. |
| |
|
| Prenatal Cell Culture Samples: | Prenatal amniotic fluid or chorionic villus samples are utilized to establish cell cultures. Six, confluent, T25 flasks of cells are required for testing. Testing is only available for FSHD1. Parental and prenatal samples are compared using OGM for 4q35 deletion detection. |
| |
|
|
|
| |
|
| Turn Around Time: | 14 days for routine FSHD1 testing or subsets of FSHD2 testing; at least 4 weeks for complete FSHD1 and FSHD2 testing; 6-8 weeks for prenatal samples, because cell cultures may require time to reach confluence or may need to be expanded. |
| |
|
| CPT Codes: |
Determination of allele size and haplotyping - 81404 Methylation testing - 81479 SMCHD1, LRIF1, and DNMT3B gene sequencing - 81479 Professional interpretation - G0452 |
| |
|
| Background: |
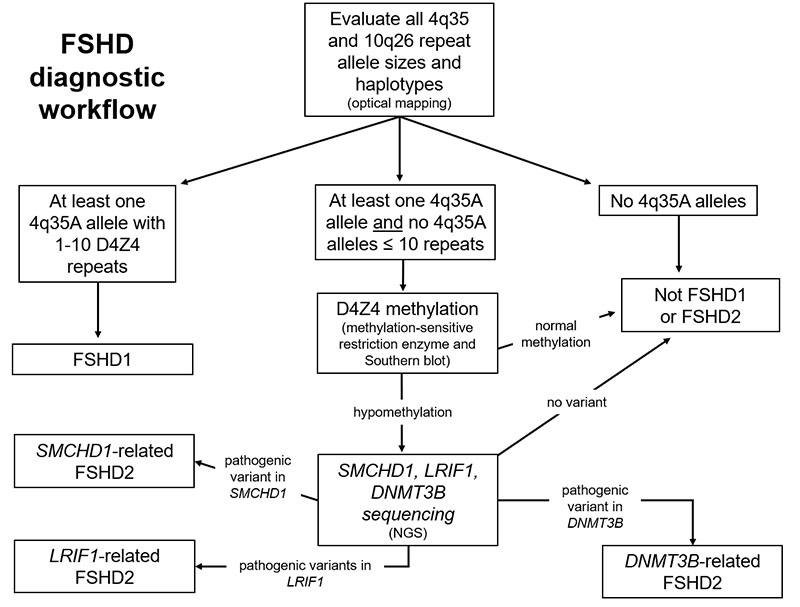
Approximately 90% of individuals affected with FSHD have a chromosome 4q35 deletion. The identification of a characteristic 4q35 deletion is more than 90% specific for the disease. Furthermore, patients with FSHD have 4qA alleles. A deleted 4q35A allele is diagnostic of FSHD type 1 (FSHD1). Approximately 5% of FSHD patients have FSHD type 2. These patients have non-deleted 4qA alleles, hypomethylation of D4Z4 repeats, and a heterozygous dominant mutation in
the SMCHD1 gene. Peripheral blood leukocytes (or cultured prenatal cells) have high molecular weight DNA extracted. FSHD analysis via Bionano DLS labeling, Saphyr instrumentation, and EnFOCUS FSHD specific pipeline/de novo assembly pipeline is based on optical mapping data collected on the Saphyr Genome Imaging instrument. Based on specific labeling and mapping of ultra-high molecular weight DNA in nanochannel arrays, optical mapping provides a high-resolution analysis of the D4Z4 repeat array. The molecules aligning to regions of interest are extracted and assembled. The resulting consensus maps are used for the Bionano EnFocus FSHD Analysis. The D4Z4 repeat regions in chromosomes 4 and 10 are sized, and the permissive and non-permissive haplotypes (4qA and 4qB, respectively) are determined. Additional structural variants and copy number gains and losses are noted in the proximity of the D4Z4 repeat array on chromosome 4 and of the SMCHD1 gene on chromosome 18. Additional testing for FSHD2 involves determining the methylation status of the D4Z4 repeats, via Southern blot analysis, that uses isolated DNA. In patients with a permissive 4qA allele and an appropriate level of hypomethylation, mutation analysis of the SMCHD1 gene is performed on a next generation sequencing platform. |
| |
|
| References: | GeneReviews - Preston MK, Tawil R, and Wang LH. Facioscapulohumeral Muscular Dystrophy |
| |
|
| Contact Information: |
Laboratory Procedures and Test Interpretation Steven A. Moore, MD, PhD 319-384-9084 or Molecular Pathology Laboratory 319-384-9568 |